rESEARCH
The primary research goal of the Morton lab is to study the fundamental mechanisms of post-transcriptional regulation of gene expression with an emphasis on RNA processing factors mutated in complex brain disorders.
Specifically, we are interested in the post-transcriptional activities of a multi-subunit RNA processing enzyme, the RNA exosome complex, in human neurodevelopment and disease. In our lab, we have taken the strategy of coupling in vivo Drosophila genetics with in vitro human iPSC-derived 2D and 3D neural induction models of RNA-mediated neurodevelopmental disorders.
Functional consequences of RNA exosome
mutations in neurological disorders

Emily Arnold
PhD Student
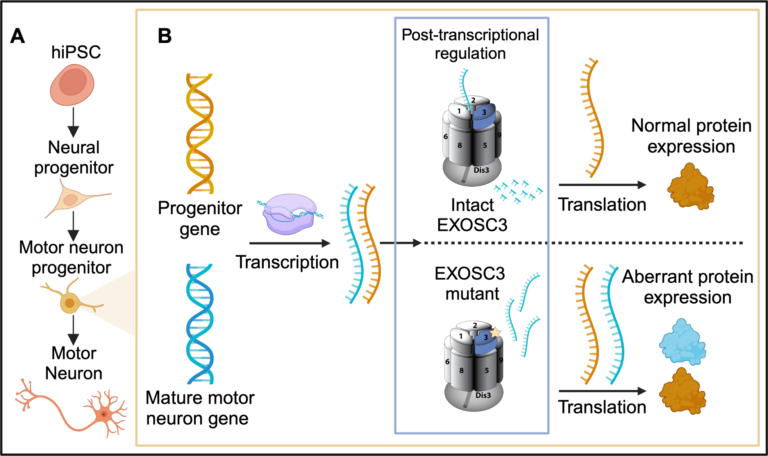
- A) Overview of the stages of directed differentiation from hiPSCs to mature phrenic spinal motor neurons.
- B) A zoomed in visualization depicting the effect of recessive missense mutations in EXOSC3 on the transcriptome and proteosome.
The diversity of cells within the brain arises from neural stem cells with specific gene expression profiles regulated by transcriptional and post-transcriptional mechanisms. While significant attention has been directed toward unraveling transcriptional activation networks driving neural stem cell differentiation and subtype specification, the role of post-transcriptional regulatory mechanisms responsible for maintaining the equilibrium between RNA production and decay to confer neuronal cell identities remains poorly understood.
Accordingly, an evolutionarily conserved RNA-regulatory protein complex, the RNA exosome, exerts critical functions via post-transcriptional processing and the destruction of RNA to control cellular differentiation. Recessive missense mutations in the EXOSC3 gene encoding a structural subunit of the RNA exosome cause Pontocerebellar Hypoplasia Type 1b (PCH1b), a severe neurodevelopmental disorder. Despite the ubiquitous and essential role of the RNA exosome in all mammalian cell types, PCH1b causes degeneration of the cerebellum, pons, and spinal motor neurons.
The tissue-specific pathology caused by mutations in the RNA exosome suggests that different tissues have varying dependencies on RNA exosome function. These observations have led to my central hypothesis that post-transcriptional regulation by the RNA exosome plays a critical role in neuronal cell-fate specification.
Towards Understanding the Molecular Basis for RNA Exosome-linked
Neurodevelopmental Disorders: A Drosophila Model

Lauryn Higginson
PhD Candidate
Post-transcriptional regulation of gene expression is critical for proper cellular development and function. Many post-transcriptional regulatory events are mediated by conserved, essential, and ubiquitously expressed RNA processing complexes, including the RNA exosome.
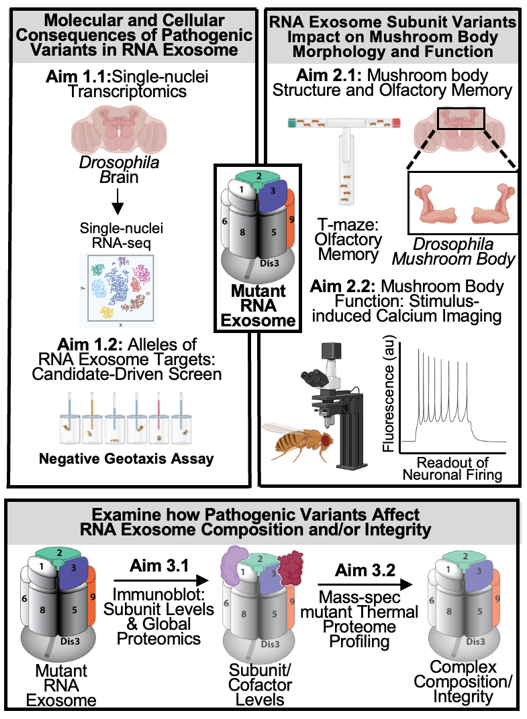
Recent clinical reports have linked recessive mutations in distinct RNA exosome subunit genes to syndromic neurodevelopmental disorders. These observations suggest an enhanced requirement for the RNA exosome within the nervous system. The goal of my research is to provide insight into how pathogenic mutations in distinct RNA exosome genes affect the function of the RNA exosome and consequently underlie neurological phenotypes.
I have engineered flies with disease mutations in different RNA exosome subunit genes to test the hypothesis that RNA exosome subunits mediate critical activities of the RNA exosome in post-transcriptional regulation of gene expression in cells of the nervous system in vivo.


Lauryn Higginson
PhD Candidate
I have engineered flies with disease mutations in different RNA exosome subunit genes to test the hypothesis that RNA exosome subunits mediate critical activities of the RNA exosome in post-transcriptional regulation of gene expression in cells of the nervous system in vivo.
I will study the functions of the RNA exosome within the fly brain to assess the molecular, cellular, and behavioral consequences of RNA exosome variants. This analysis will provide insight into tissue-specific functions of RNA exosome within the nervous system in vivo.
Investigating the Molecular and Cellular Basis of RNA Exosome-linked Pontocerebellar Hypoplasia Type1b in Human 3D Brain Organoids

Nina Barr
PhD Candidate

The diversity of cells within the brain is derived from neuronal stem cells with specific gene expressions controlled by transcriptional and post-transcriptional regulatory mechanisms. How post-transcriptional mechanisms accurately balance RNA production and decay to confer neuronal cell identities during differentiation is poorly understood.
My project aims to understand the role of a post-transcriptional RNA-regulatory complex, the RNA exosome, in shaping neuronal cell diversity and its connection to a severe neurodevelopmental disorder (NDD), Pontocerebellar Hypoplasia Type 1b (PCH1b), clinically characterized by atrophy of the cerebellum and pons. Recessive missense mutations in the RNA exosome subunit gene, EXOSC3, cause PCH1b.
Given that the RNA exosome is an essential and ubiquitously expressed ribonuclease complex, these observations support the idea that the RNA exosome has enhanced functions within the nervous system. The central hypothesis underlying the proposed work is that the RNA exosome processes specific target RNAs essential for proper progenitor proliferation and neuronal differentiation within the brain. We have generated human induced pluripotent stem cells (hiPSCs) modeling two distinct PCH1b disease-causing EXOSC3 mutations linked to moderate or severe PCH1b disease and isogenic controls via CRISPR engineering to reveal the molecular and cellular basis of PCH1b.











